Section: New Results
Computational Anatomy
Inconsistency of Template Estimation in Quotient Spaces
Participants : Loïc Devilliers [Correspondent] , Stéphanie Allassonnière [Université Paris-Descartes] , Alain Trouvé [ENS Paris-Saclay] , Xavier Pennec.
Inria postdoctoral fellowship for 16 months
Template estimation, Fréchet Mean, Quotient Spaces, Inconsistency, Consistency Bias
-
A central issue in Computational Anatomy is to compute an unbiased template prototype of our data images (the template) in the presence of two effects: the noise in the ambient space and the unknown registration of the data. The template estimation is usually performed by minimizing the discrepancy after registration (and iterating), which corresponds geometrically to the computation of the Fréchet mean in the quotient space. So far, it was generally believed that the template estimation with this method was unbiased.
-
We show in this work that inconsistency is in fact the general situation when the ambient space is an infinite dimensional linear space. In [15] we prove that this method is generally inconsistent when the action is isometric. Moreover the consistency bias has been quantified [35] thanks to a Taylor expansion in the noise level. Besides, we provide proofs of inconsistency for non isometric action [15] when the noise level is large enough.
Geometric statistics for Computational Anatomy
Participants : Nina Miolane [Correspondent] , Xavier Pennec.
This work is conducted jointly with the Department of Statistics of Stanford, in the context of the associated team GeomStats of the program Inria@SiliconValley.
Statistics, Computational Anatomy, Differential Geometry, Template shape, asymptotic bias
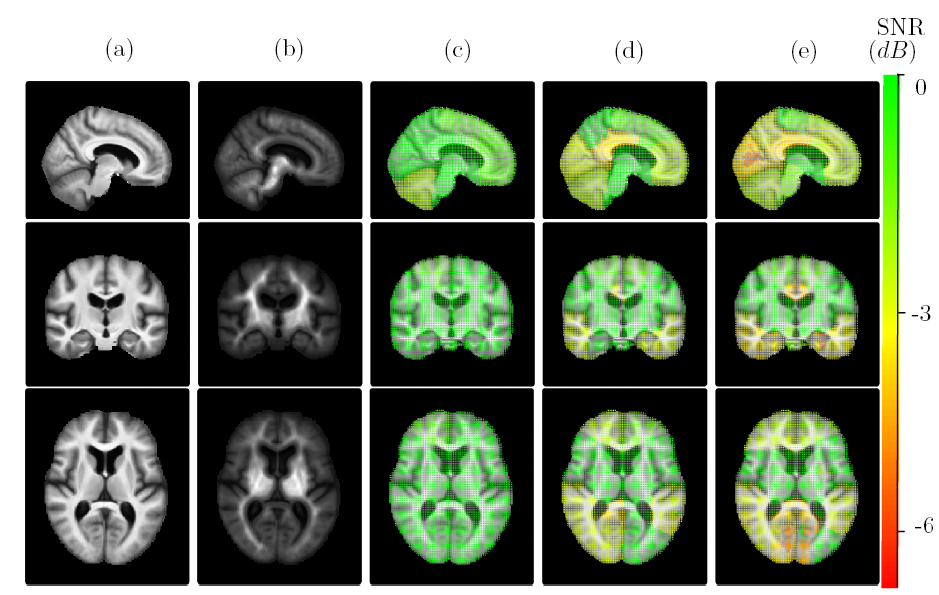
The usual algorithm of brain template estimation is asymptotically biased, therefore inconsistent: even with an infinite number of brain images in the database, the template estimate may not converge to the brain anatomy it is meant to estimate. In [22]:
-
we present a methodology that quantifies spatially the brain template's asymptotic bias, see Figure 14,
-
we propose a topologically constrained adaptation of the template computation, that constructs a hierarchical template with bounded bias, and we apply it to the Open Access Series of Imaging Studies (OASIS) database.
|
SVF-Net: Learning Deformable Registration Using Shape Matching
Participants : Marc Michel Rohe [Correspondent] , Xavier Pennec, Maxime Sermesant.
The authors acknowledge the partial funding by the EU FP7-funded project MD-Paedigree (Grant Agreement 600932).
Registration, Deep Learning, Shape Matching
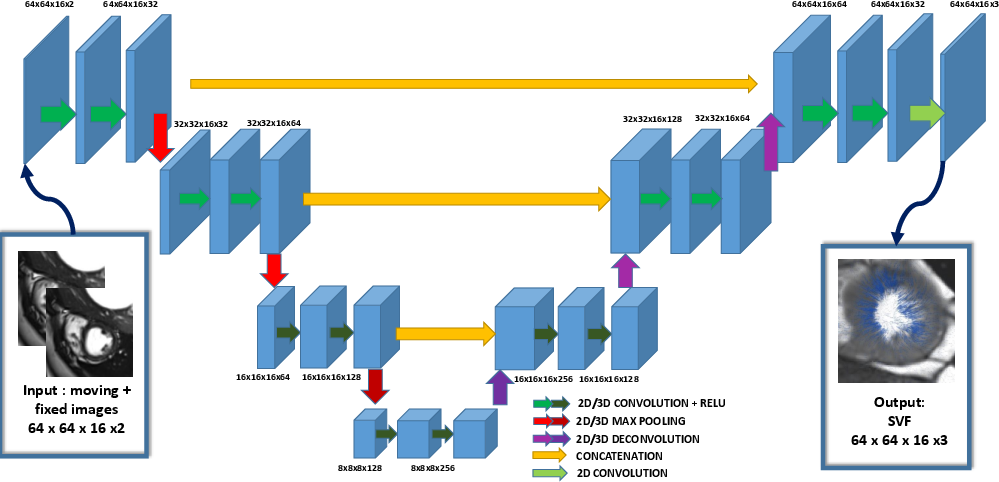
We propose an innovative approach for registration based on the deterministic prediction of the parameters from both images instead of the optimization of a energy criteria [44]. The method relies on a fully convolutional network (see Fig. 15). Whereas convolutional networks have seen a widespread expansion and have been already applied to many medical imaging problems such as segmentation and classification, its application to registration has so far faced the challenge of defining ground truth data on which to train the algorithm. Here, we present a novel training strategy to build reference deformations which rely on the registration of segmented regions of interest. The speed and robustness of this registration algorithm make it a strong candidate within a multi-atlas segmentation pipeline [45].
|
Reduced Representation of Segmentation and Tracking in Cardiac Images for Group-Wise Longitudinal Analysis
Participants : Marc Michel Rohe [Correspondant] , Xavier Pennec, Maxime Sermesant.
The authors acknowledge the partial funding by the EU FP7-funded project MD-Paedigree (Grant Agreement 600932).
Medical image analysis, Non-rigid registration, Deep learning, Statistical model reduction, Longitudinal analysis
We study image-based methods for the analysis of cardiac motion to enable group-wise statistics, automatic diagnosis and longitudinal study [10]. This is achieved by combining advanced medical image processing with machine learning methods and statistical modelling. The first axis of this work is to define an automatic method for the segmentation of the myocardium. The second axis of this work is focused on the improvement of cardiac motion tracking methods in order to define relevant low-dimensional representations. Finally, in the last axis, we apply the previously defined representation to the problem of diagnosis and longitudinal analysis. These three axes form an end to end framework for the study of cardiac motion starting from the acquisition of the medical images to their automatic analysis. Such a framework could be used for diagnosis and therapy planning in order to improve the clinical decision making with a more personalised computer-aided medicine.
A model of brain morphological evolution
Participants : Raphaël Sivera [Correspondent] , Hervé Delingette, Marco Lorenzi, Xavier Pennec, Nicholas Ayache.
Longitudinal modeling, deformation framework, brain morphology, Alzheimer's disease, aging.
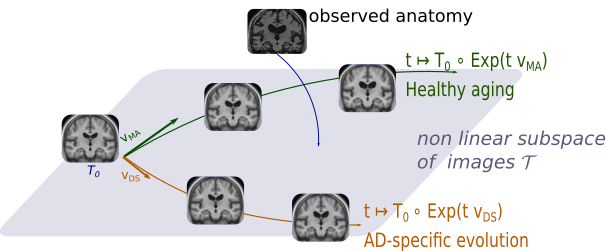
We proposed a deformation-based generative model of the brain morphological evolution that can joinly describes the effect of aging and Alzheimer's disease. It relies on longitudinal description of the aging and disease consequences and can be use to compute image-based cross-sectional progression markers (see Figure 16). This approach is able to propose a description of the disease evolution, population and subject-wise.
|
Statistical Learning of Heterogeneous Data in Large-Scale Clinical Databases
Participants : Clement Abi Nader [Correspondent] , Nicholas Ayache, Marco Lorenzi.
The research takes place within the MNC3 initiative (Médecine Numérique: Cerveau, Cognition, Comportement) funded by Université Côte d’Azur (UCA), and is performed in collaboration with the Institut Claude Pompidou (CHU of Nice).
Longitudinal modeling, brain structure, Alzheimer's disease, aging, Gaussian processes, ICA.
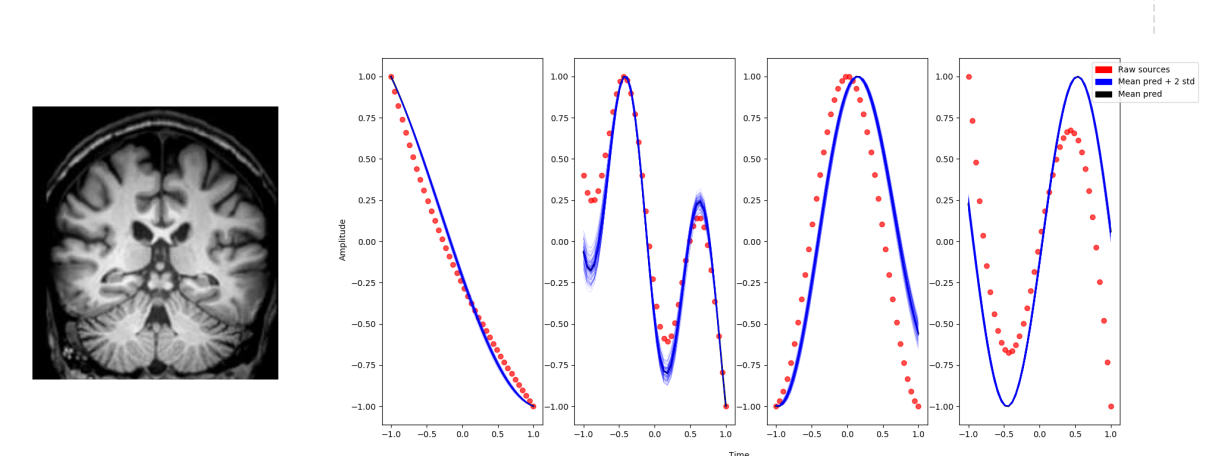
Through this project we aim at developing novel scalable spatio-temporal analysis tools to identify clinical and biological modulators of structural and functional brain changes across time. The project relies on the extension of current un-/semi-supervised image analysis approaches (such as independent component analysis, ICA) to encode priors on spatial and temporal properties of the signal measured in brain images. The application to currently available large-scale biomedical datasets (such as the UK Biobank) will be addressed by focusing on scalable and distributed learning methods.
|